Abstract
Background: Viral myocarditis (VMC) is a common cardiovascular disease, and circular RNAs (circRNAs) have been identified to play an important role in the pathophysiology of cardiovascular disease. However, the clinical significance, biological functions, and regulatory mechanisms of circRNAs in VMC remain poorly understood. Therefore, this study explored the biological functions and regulatory mechanisms of circ-ACSL1 in VMC.
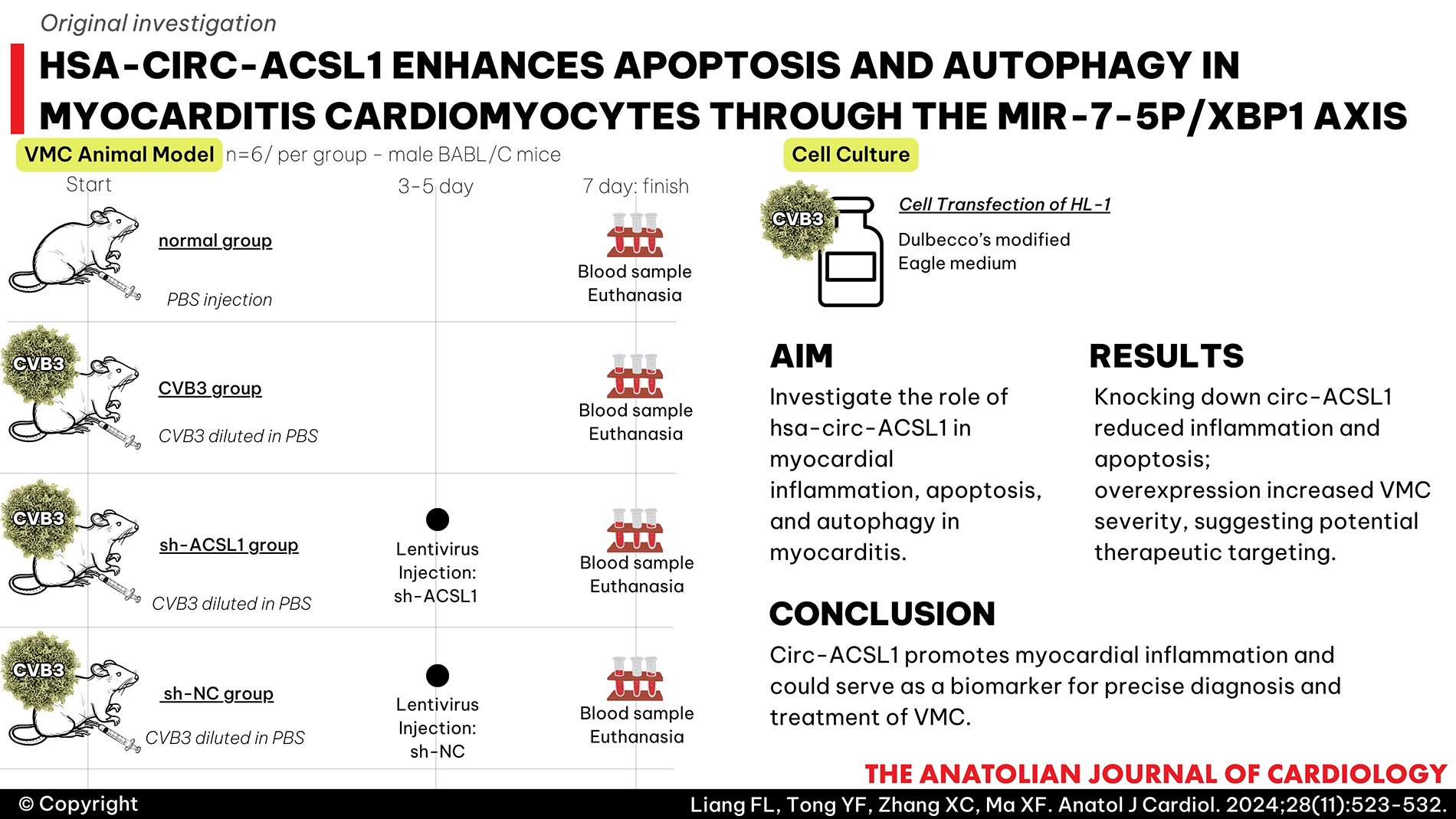
Methods: The animal and cell models of VMC were established by infecting BABL/C mice and interleukin-2 cells with coxsackievirus B3 (CVB3). Pro-inflammatory factors, markers of myocardial injury, apoptosis, and autophagy were detected to evaluate the degree of myocardial inflammation and myocardial injury after altering circ-ACSL1, microRNA-7-5p (miR-7-5p), and X-box binding protein 1 (XBP1) expression alone or in combination.
Results: Knocking down circ-ACSL1 could inhibit inflammation, autophagy, and apoptosis in VMC animals and cells. Mechanistically, circ-ACSL1 targeted miR-7-5p to regulate the downstream target XBP1. In addition, depleting miR-7-5p rescued the therapeutic effect of depleting circ-ACSL1. Overexpression of circ-ACSL1 aggravated VMC; however, this effect was saved by knocking down XBP1.
Conclusion: By competitively absorbing miR-7-5p, circ-ACSL1 increases XBP1 expression and aggravates myocardial inflammation. Meaningfully, VMC treatment may benefit from circ-ACSL1 as a potential biomarker for precise diagnosis and as a potential therapeutic target.
Graphical Abstract
Highlights
- Knocking down circ-ACSL1 inhibits inflammation, apoptosis, and autophagy of VMC cardiomyocytes.
- miR-7-5p downregulation can reduce the therapeutic effect of depleting circ-ACSL1 on VMC.
- circ-ACSL1 overexpression promotes VMC, but this effect is saved by depleting XBP1.
Introduction
Myocarditis (MC) is an inflammatory disease that involves the myocardium and leads to cardiomyopathy. It is caused by a variety of causes such as immune damage, bacterial infection, viral infection, etc.,1 and is a potentially life-threatening myocardial inflammatory disease.2 Viral infection is the most common type of MC, and coxsackievirus B3 (CVB3), a cardiac virus, is the most common cause of MC.3 In some cases, viral MC (VMC) can progress to chronic MC, dilated cardiomyopathy, and congestive heart failure, which may require a heart transplant.4
The autophagic process involves the degradation and recycling of unwanted proteins, organelles, and other cellular components.5 The process of autophagy is generally seen as a protective response to stress.6 Viruses can utilize autophagy to evade degradation and facilitate viral replication and acquisition of metabolites, while over-activated autophagy also promotes apoptosis and inflammatory cascades.7 In the body, apoptosis occurs naturally, regulating cell death as part of homeostasis. As cell survival is determined by the balance between autophagy and apoptosis, autophagy has a complex relationship with apoptosis.8
The discovery and exploration of circular RNAs (circRNAs) date back decades.9 It has been reported that circRNAs are involved in many pathophysiological processes due to various biological functions.10 In addition, circRNA is generally low in expression and often exhibit cell type and tissue specificity.11 circ-ACSL1 shows up-regulated expression in MC and has pro-inflammatory properties in MC.12 Moreover, hsa-circ-ACSL1 may constitute as a potential biomarker for fulminant MC.13 However, circ-ACSL1 is not well-explored from a biological perspective in MC. In terms of mechanism, emerging reports support that circRNA can act as a competing endogenous RNA to regulate messenger RNA (mRNA) expression via spongy micro RNAs (miRNAs).14-
miR-7-5p, a type of microRNA, has been shown to be regulated by circRNAs.18 microRNA-7 is reported to be upregulated in ischemia/reperfusion (I/R) injury.19,
Based on this, the present study intends to investigate the mechanism of action of circ-ACSL1 involved in the pathogenesis and progression of VMC and promotion of apoptosis and autophagy in MC cardiomyocytes through circRNA–miRNA–mRNA network. Elucidation of the potential molecular mechanisms mediated by circACSL1 may lead to the development of promising therapeutic candidates for VMC.
Methods
Laboratory Animals and Viruses
Thirty male BABL/C mice (4-6 weeks, 16-19 g) were purchased from SLAC Laboratory Animal Co., Ltd. (Shanghai, China). A standard diet and water were provided to the mice for a week, and they were kept at 25 ± 2°C with 60-70% humidity and a 12-hour light/dark cycle.
CVB3 was obtained (ATCC, USA), and HL-1 cells (ATCC) were used as vectors for CVB3 replication. The virus titer of CVB3 was determined using the tissue culture infectious dose 50 (TCID50) method.
VMC Animal Model
BABL/C mice were randomly divided into normal group, CVB3 group, sh-ACSL1 group, and sh-NC group (6 mice per group). In the normal group, 100 μL phosphate buffered saline (PBS) was injected intraperitoneally, while 100 μL CVB3 diluted in PBS (103 TCID50) was injected intraperitoneally in the other 3 groups. Body weight and death were recorded daily. Seven days after injection, blood samples were collected from the eyeball, and serum was separated by centrifugation at 3000 ×
Lentivirus Injection
The mice were injected with 100 μL of circ-ACSL1 shRNA lentiviral vector (sh-ACSL1) or sh-NC (1 × 108 TU/mL) through the tail vein on days 3 and 5 after CVB3 infection. The lentiviral vectors were synthesized by GenePharma (Shanghai, China).
Terminal Deoxynucleotidyl Transferase dUTP Nick-End Labeling (TUNEL) Staining
Myocardial tissues fixed in 10% paraformaldehyde were sectioned to 4 μm after paraffin embedding. Tissue paraffin sections were washed with xylene for 5 minutes and treated with 100%, 95%, 90%, 80%, and 70% ethanol, each for 3 minutes. The myocardial tissues were then treated with protease K for 30 minutes, soaked in a blocking buffer for 10 minutes, and added with a mixture of TdT (2 μL) and green fluorescent fluorescein isothiocyanate (FITC) probe-labeled dUTP (48 μL) (Beyotime, Shanghai, China) for 1 hour. The tissues were sealed with an anti-fluorescence quencher and observed under a fluorescence microscope. Nuclei were stained with DAPI. TUNEL positive cells (%) = number of green cells/total cells × 100%.
Enzyme-Linked Immunosorbent Assay (ELISA)
Creatine Kinase Isoenzymes (CK-MB), Cardiac troponin I (cTnI), and B-type natriuretic peptide (BNP) in serum and cells were measured by Mouse CK-MB ELISA Kit, Mouse TNNI3/cTn-I ELISA Kit, and BNP ELISA Kit, respectively. Interleukin-6 (IL-6), interleukin-1β (IL-β), and tumor necrosis factor-α (TNF-α) in myocardial tissue and cell culture supernatant were determined by ELISA Kits. All the above kits were purchased from Sangon (Shanghai, China). Optical density was measured using a microplate reader (EL 340, Bio-Tek Instruments, USA).
Western Blot
Myocardial tissue and cells were separately lysed in radioimmunoprecipitation lysis buffer (Beyotime). Proteins that were extracted were quantified by a bicinchoninic acid kit (Beyotime). Proteins were mixed with a sample buffer (Beyotime) and treated in a boiling water bath for 3 minutes. After electrophoresis at 80 V for 30 minutes, the separation continued at 120 V for 1-2 hours. The proteins were transferred to a membrane using a 300 mA ice bath for 60 minutes and soaked in a blocking buffer for 60 minutes at room temperature or overnight at 4°C. The protein was combined with anti-β-actin (ab8227, 1 : 1000), LC3B (ab51520, 1 : 3000), Beclin-1 (ab210498, 1 : 1000), Bcl-2 (ab182858, 1 : 2000), Bax (ab182733, 1 : 2000), Cleaved caspase 3 (ab2302, 1 : 500), XBP1 (ab37152, 1 μg/mL), p-p65 (ab86299, 1 : 2000), and p65 (ab16502, 1 : 1000, all from Abcam, UK), and transferred to the secondary antibody (Goat Anti-Rabbit immunoglobulin G, ab205718, 1 : 2000). A developer was added to the membrane, and protein bands were detected using a chemiluminescent imaging system (Gel Doc XR, Bio-rad).
Cell Culture
HL-1 cells were placed in Dulbecco’s modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Gibco, USA) and 1% penicillin–streptomycin (HyClone, USA) at 37°C with 5% CO2.
HL-1 cells and CVB3 (100 TCID50) were incubated in a serum-free medium for 2 hours, followed by incubation in a normal medium.
Cell Transfection
circ-ACSL1 and XBP1 were overexpressed using pcDNA3.1 (Invitrogen, Thermo Fisher Scientific). Small interfering RNAs targeting circ-ACSL1 and XBP1 (si-circ-ACSL1 and si-XBP1), as well as negative control siRNAs (si-circ-ACSL1-NC, si-XBP1-NC), miR-7-5p-mimic, miR-7-5p-inhibitor, mimic-NC, and inhibitor-NC, were synthesized by GenePharma. Using Lipofectamine 3000 (Invitrogen), these oligonucleotides and plasmids were transfected into HL-1 cells, and the culture medium was replaced 6 hours later. After 48 hours, transfection efficiency was evaluated using real-time reverse transcriptase-polymerase chain reaction (RT-qPCR) or Western blot.
Flow Cytometry
Apoptotic cells were measured using the FITC annexin V apoptosis assay kit I (BD Biosciences, USA). A cell suspension (3 mL, 1 × 105/mL) was centrifuged at 500 r/min for 5 minutes, and the culture medium was discarded after centrifugation. The remaining pellet was centrifuged at 500 r/min for 5 minutes, and the resulting cells were resuspended in 100 μL binding buffer with 5 μL annexin V-FITC and 5 μL propidium iodide (PI). After 15 minutes, FITC and PI fluorescence were detected by flow cytometry.
Monodansylcadaverine Staining
Autophagy was observed around the positive nuclei, and all acidic vacuoles were stained. Cell slides were prepared overnight and incubated with 0.05 mmol/L monodansylcadaverine (MDC) (Huzheng, Shanghai, China) in a water bath at 37℃ for 15 minutes. After fixation with 4% paraformaldehyde for 15 minutes, anti-fluorescence quenched slides were observed by fluorescence microscopy in the dark.
RNAse R Test
Total RNA isolation of HL-1 cells was performed with TRIzol reagent (Invitrogen). Then, 2 μg RNA was incubated with 5 U/μg RNAse R (Epicentre Technologies, USA) at 37℃ for 24 hours. Linear RNA and circular RNA were analyzed by RT-qPCR.
Actinomycetes D Test
A total RNA sample was extracted from HL-1 cells after exposure to 100 ng/mL actinomycin D (Merck, Germany) at 0, 4, 8, 12, and 24 hours for RT-qPCR analysis of circ-ACSL1 stability.
Reverse Transcriptase-Polymerase Chain Reaction
Total RNA was extracted from cardiomyocytes and tissues using SparkZol reagent (SparkJade, China). The quantity and quality of total RNA were estimated with a NanoDrop ND-2000 spectrophotometer (NanoDrop, USA). Complementary DNA (cDNA) was synthesized using Evo M-MLV RT Premix (AG 11706, ACCURATE BIOTECHNOLOGY, China) and Mir-XTM miRNA first chain synthesis Kit (Takara, Dalian, China). RT-qPCR was performed with the SYBR Green Premix Pro Taq HS qPCR Kit (AG 11701, ACCURATE BIOTECHNOLOGY). Primers are shown in
RNA Pull Down
Using the TranscriptAid T7 high-yield transcription Kit (Thermo Fisher Scientific), miR-7-5p wild type (WT) and mutant (Mut) miR-7-5p were transcribed. Biotinized miR-7-5p-WT or miR-7-5p-Mut were incubated with cell lysate at 48°C for 1 hour. After purification, RNA was eluted with elution buffer, and RT-qPCR was performed to check circ-ACSL1 and XBP1 expression.
Luciferase Reporter Gene Assay
Potential binding sequences of miR-7-5p to circ-ACSL1 and XBP1 3′-UTR were predicted by starBase database (
Statistical Analysis
Statistical analysis was carried out using SPSS v.24.0 software, and visualization of data was obtained by Graphpad prism 8.0 software. Results were obtained from at least 3 independent replicated experiments. The normality of data distribution was assessed in this study using the Shapiro–Wilk test, and data were expressed as mean ± SD. The 2-tailed Student’s
Results
High Expression and Stability of Circ-ACSL1 in VMC
The volcano map of differential expression and function of circRNAs in children with fulminant MC13 showed that circ-ACSL1 was up-regulated (
After CVB3 injection, mice injected with CVB3 continued to lose weight from day 3 (
After RNAse R digestion and actinomycin D treatment, circACSLl in HL-1 cells was detected by RT-qPCR to verify the closed-loop and stability of circACSLl. circ-ACSL1 was resistant to RNAse compared to ACSL1, suggesting that circ-ACSL1 is a circRNA (
Knocking Down circ-ACSL1 Inhibits Inflammation, Apoptosis, and Autophagy of VMC Cardiomyocytes
shRNA lentiviral vectors targeting circ-ACSL1 were injected into VMC mice. The interference RNA si-circ-ACSL1 was transfected into infected HL-1 cells. Then, RT-qPCR confirmed that circ-ACSL1 expression in myocardial tissue and HL-1 cells decreased with the depleting circ-ACSL1 (
Circ-ACSL1 Targets miR-7-5p Adsorption
RT-qPCR detected low expression of miR-7-5p in myocardial tissue of VMC mice and CVB3-treated HL-1 cells (
miR-7-5p Downregulation can Reduce the Therapeutic Effect of Depleting Circ-ACSL1 on VMC
HL-1 cells were transfected with si-circ-ACSL1 + miR-7-5p inhibitor. RT-qPCR results showed that si-circ-ACSL1 decreased circ-ACSL1 and increased miR-7-5p levels, while the miR-7-5p inhibitor saved the expression increase of miR-7-5p (
miR-7-5p Targets XBP1
High expression of XBP1 was detected by RT-qPCR and Western bot in VMC mice and CVB3-treated HL-1 cells (
Circ-ACSL1 Overexpression Promotes VMC, But This Effect is Saved by Depleting XBP1
HL-1 cells were transfected with pcDNA-circ-ACSL1, pcDNA-circ-ACSL1 + si-XBP1, and pcDNA, respectively. RT-qPCR results confirmed that pcDNA-circ-ACSL1 in HL-1 cells enhanced circ-ACSL1 and XBP1 expression, while si-XBP1 saved XBP1 expression (
Discussion
Viral myocarditis (VMC) is an inflammatory disease resulting from a viral infection, causing immunological responses resulting in dysfunction and impaired contractility in the heart.23 The molecular regulation of VMC progression remains an elusive problem. There has been a growing body of evidence that circRNAs are crucial regulators of cardiovascular diseases.24,
Circular RNAs often act as molecular sponges for miRNAs to indirectly regulate gene expression.27 In this study, circACSL 1 contains a conserved miR-7-5p target site, which was validated by luciferase reporter gene assay in the HL-1in vitro cell model. A number of miRNAs have been reported to exhibit aberrant expression levels in VMC, and a growing body of evidence has highlighted the role of miRNAs in MC pathogenesis.28-
XBP1 expression is increased, and XIAP expression is decreased in LPS-induced H9c2 cells, and XBP1 aggravates LPS-induced cardiomyocyte injury by down-regulating XIAP through activating the NFκB signaling pathway.32 Similar to this result, in our study, XBP1 was found to be highly expressed in CVB3-infected myocardial tissues and cardiomyocytes, and XBP1 was considered as a target gene of miR-7-5p, which showed a negative regulatory relationship with miR-7-5p expression. In addition, it can be positively regulated by circACSL1. Knockdown of XBP 1 was able to rescue the promotion of myocardial inflammation, apoptosis, and autophagy by overexpression of circACSL1.
In conclusion, the present study identified that circACSL1 was significantly upregulated in VMC and significantly exacerbated inflammation in VMC, resulting in increased apoptotic cell death and increased cellular autophagy. The mechanism by which this occurs may be that circACSL1 regulates XBP1 expression through targeting adsorption of miR-7-5p.
Footnotes
References
- Daba TM, Zhao Y, Pan Z. Advancement of mechanisms of Coxsackie virus B3-induced myocarditis pathogenesis and the potential therapeutic targets. Curr Drug Targets. 2019;20(14):1461-1473. https://doi.org/10.2174/1389450120666190618124722
- Fung G, Luo H, Qiu Y, Yang D, McManus B. Myocarditis. Circ Res. 2016;118(3):496-514. https://doi.org/10.1161/CIRCRESAHA.115.306573
- Garmaroudi FS, Marchant D, Hendry R. Coxsackievirus B3 replication and pathogenesis. Future Microbiol. 2015;10(4):629-653. https://doi.org/10.2217/fmb.15.5
- Liu YL, Wu W, Xue Y. MicroRNA-21 and -146b are involved in the pathogenesis of murine viral myocarditis by regulating TH-17 differentiation. Arch Virol. 2013;158(9):1953-1963. https://doi.org/10.1007/s00705-013-1695-6
- Wang X, Dai Y, Ding Z, Khaidakov M, Mercanti F, Mehta JL. Regulation of autophagy and apoptosis in response to angiotensin II in HL-1 cardiomyocytes. Biochem Biophys Res Commun. 2013;440(4):696-700. https://doi.org/10.1016/j.bbrc.2013.09.131
- Yoshida GJ. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: from pathophysiology to treatment. J Hematol Oncol. 2017;10(1):67-. https://doi.org/10.1186/s13045-017-0436-9
- Meng Y, Sun T, Wu C, Dong C, Xiong S. Calpain regulates CVB3 induced viral myocarditis by promoting autophagic flux upon infection. Microbes Infect. 2020;22(1):46-54. https://doi.org/10.1016/j.micinf.2019.07.001
- Wang X, Guo Z, Ding Z, Mehta JL. Inflammation, autophagy, and apoptosis after myocardial infarction. J Am Heart Assoc. 2018;7(9):-. https://doi.org/10.1161/JAHA.117.008024
- Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1-7. https://doi.org/10.1007/s12282-017-0793-9
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675-691. https://doi.org/10.1038/s41576-019-0158-7
- Wilusz JE, Sharp PA. Molecular biology. a circuitous route to noncoding RNA. Science. 2013;340(6131):440-441. https://doi.org/10.1126/science.1238522
- Zhang L, Han B, Liu H. Circular RNA circACSL1 aggravated myocardial inflammation and myocardial injury by sponging miR-8055 and regulating MAPK14 expression. Cell Death Dis. 2021;12(5):487-. https://doi.org/10.1038/s41419-021-03777-7
- Zhang L, Han B, Wang J. Differential expression profiles and functional analysis of circular RNAs in children with fulminant myocarditis. Epigenomics. 2019;11(10):1129-1141. https://doi.org/10.2217/epi-2019-0101
- Jin X, Feng CY, Xiang Z, Chen YP, Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7(41):66455-66467. https://doi.org/10.18632/oncotarget.12186
- Rong D, Sun H, Li Z. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271-73281. https://doi.org/10.18632/oncotarget.19154
- Yuan W, Zhou R, Wang J. Circular RNA Cdr1as sensitizes bladder cancer to cisplatin by upregulating APAF1 expression through miR-1270 inhibition. Mol Oncol. 2019;13(7):1559-1576. https://doi.org/10.1002/1878-0261.12523
- Zhou J, Li L, Hu H. Circ-HIPK2 accelerates cell apoptosis and autophagy in myocardial oxidative injury by sponging miR-485-5p and targeting ATG101. J Cardiovasc Pharmacol. 2020;76(4):427-436. https://doi.org/10.1097/FJC.0000000000000879
- Zhu M, Li Y, Liu L, Zhai X. Circ_0057452 sponges miR-7-5p to promote keloid progression through upregulating GAB1. Cell Cycle. 2022;21(23):2471-2483. https://doi.org/10.1080/15384101.2022.2102796
- Ren XP, Wu J, Wang X. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119(17):2357-2366. https://doi.org/10.1161/CIRCULATIONAHA.108.814145
- Li B, Li R, Zhang C. MicroRNA-7a/b protects against cardiac myocyte injury in ischemia/reperfusion by targeting poly(ADP-ribose) polymerase. PLoS One. 2014;9(3):e90096-. https://doi.org/10.1371/journal.pone.0090096
- Gupta S, Deepti A, Deegan S, Lisbona F, Hetz C, Samali A. HSP72 protects cells from ER stress-induced apoptosis via enhancement of IRE1alpha-XBP1 signaling through a physical interaction. PLoS Biol. 2010;8(7):e1000410-. https://doi.org/10.1371/journal.pbio.1000410
- Xu W, Wang C, Hua J. X-box binding protein 1 (XBP1) function in diseases. Cell Biol Int. 2021;45(4):731-739. https://doi.org/10.1002/cbin.11533
- Li B, Xie X. A20 (TNFAIP3) alleviates viral myocarditis through ADAR1/miR-1a-3p-dependent regulation. BMC Cardiovasc Disord. 2022;22(1):10-. https://doi.org/10.1186/s12872-021-02438-z
- Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588-5600. https://doi.org/10.1002/jcp.27384
- Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in cardiovascular disease: an overview. BioMed Res Int. 2017;2017():5135781-. https://doi.org/10.1155/2017/5135781
- Fan S, Hu K, Zhang D, Liu F. Interference of circRNA HIPK3 alleviates cardiac dysfunction in lipopolysaccharide-induced mice models and apoptosis in H9C2 cardiomyocytes. Ann Transl Med. 2020;8(18):1147-. https://doi.org/10.21037/atm-20-5306
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344-352. https://doi.org/10.1038/nature12986
- Corsten MF, Heggermont W, Papageorgiou AP. The microRNA-221/-222 cluster balances the antiviral and inflammatory response in viral myocarditis. Eur Heart J. 2015;36(42):2909-2919. https://doi.org/10.1093/eurheartj/ehv321
- Wang J, Han B. Dysregulated CD4+ T cells and microRNAs in myocarditis. Front Immunol. 2020;11():539-. https://doi.org/10.3389/fimmu.2020.00539
- Goldberg L, Tirosh-Wagner T, Vardi A. Circulating microRNAs: a potential biomarker for cardiac damage, inflammatory response, and left ventricular function recovery in pediatric viral myocarditis. J Cardiovasc Transl Res. 2018;11(4):319-328. https://doi.org/10.1007/s12265-018-9814-0
- Deng B, Zheng X, Zheng X, Tian L, Zhang Y. Propofol inactivates NF-κB pathway to inhibit lipopolysaccharide-induced myocarditis via miR-142-5p/SOCS1 axis. J Biol Regul Homeost Agents. 2023;37(6):2927-2934.
- Zhang C, Chen X, Wang C, Ran Y, Sheng K. Inhibition of XBP1 alleviates LPS-induced cardiomyocytes injury by upregulating XIAP through suppressing the NF-kappaB signaling pathway. Inflammation. 2021;44(3):974-984. https://doi.org/10.1007/s10753-020-01392-w


