2Clinical Laboratory, Tangshan Fengnan District Hospital, Hebei, China
3Institute of Drug Inspection and Testing, Tangshan Food and Drug Comprehensive Inspection and Testing Center, Hebei, China
4Department of Reproductive Genetics, Tangshan Maternal and Children Health Hospital, Hebei, China
5Department of Nursing, Tangshan Gongren Hospital, Hebei, China
6Obstetrics and Gynecology Department, Tangshan Maternal and Children Health Hospital, Hebei, China
7Department of Breast Surgery, Tangshan Central Hospital, Hebei, China
Abstract
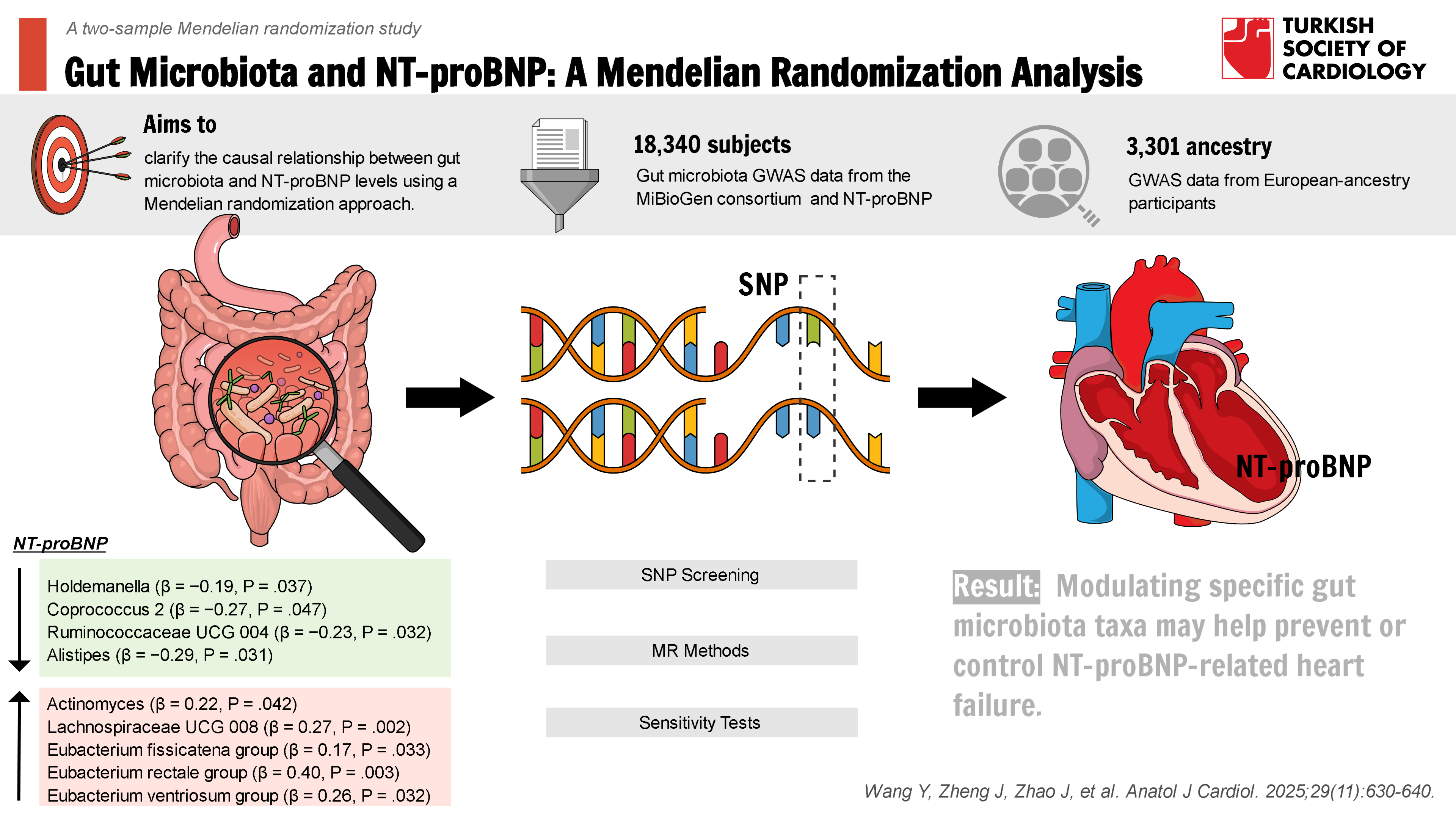
Background: This study aimed to clarify the potential causal relationship between gut microbiota (GM) and N-terminal pro-B-type natriuretic peptide (NT-proBNP) using Mendelian randomization (MR) analysis.
Methods: Genome-wide association study of intestinal flora and NT-proBNP was conducted, and the instrumental variables (IVs) were screened out to assess the causal association between intestinal flora and NT-proBNP. The 2-sample MR analysis was performed using the inverse variance weighted (IVW), MR-Egger, weighted model and simple model methods, respectively, to assess the causal association between intestinal flora and NT-proBNP using the odds ratio. Sensitivity analyses were also performed using the leave-one-out method and MR-Egger intercept test. The MR-PRESSO global test was used to detect horizontal pleiotropy, and Cochran Q was used to detect heterogeneity. Finally, forest plots, scatter plots, and funnel plots of the IVs were generated.
Results: Based on the IVW method, a total of 9 out of 104 GM genera were identified as causally associated with NT-proBNP levels (P < .05). The genera Holdemanella (β = −0.19, 95% CI: −0.36 to −0.01, P = .037), Coprococcus 2 (β = −0.27, 95% CI: −0.54 to 0.00, P = .047), Ruminococcaceae UCG 004 (β = −0.23, 95% CI: −0.45 to −0.02, P = .032), and Alistipes (β = −0.29, 95% CI: −0.55 to −0.03, P = .031) were negatively associated with NT-proBNP; genera Actinomyces (β = 0.22, 95% CI: 0.01-0.44, P = .042), Lachnospiraceae UCG 008 (β = 0.27, 95% CI: 0.1-0.44, P = .002), Eubacterium fissicatena group (β = 0.17, 95%CI: 0.01-0.32, P = .033), Eubacterium rectale group (β = 0.40, 95% CI: 0.13-0.67, P = .003), and Eubacterium ventriosum group (β = 0.26, 95% CI: 0.02-0.49, P = .032) were positively associated with NT-proBNP. In addition, the results of the sensitivity analysis of the leave-one-out method were stable, there was no horizontal multiplicity in the MR-Egger intercept test and the MR-PRESSO global test, and there was no heterogeneity in the Cochran Q-test.
Conclusions: The study found the causal relationship between GM and NT-proBNP. As a clinical predictor of heart failure, NT-proBNP levels could potentially be modulated through clinical interventions involving GM, further reducing the risk of heart failure.
Graphical Abstract
Highlights
- Causal link between gut microbiota (GM) and NT-proBNP: Certain bacteria like are negatively associated with NT-proBNP, potentially aiding heart failure prevention.
- Potential for GM regulation: Modulating GM could lower NT-proBNP levels and reduce heart failure risk.
- Robust results: Sensitivity analysis confirmed the results’ reliability and the validity of the causal relationship.
Introduction
N-terminal pro-B-type natriuretic peptide (NT-proBNP), a derivative of the precursor molecule of B-type natriuretic peptide (BNP), serves as a pivotal biomarker for evaluating cardiovascular function and assessing the risk of cardiac disease, particularly heart failure1 Li-natriuretic peptide-guided therapy improves cardiovascular outcomes of patients with high-risk heart failure and reduced ejection fraction disease.2 In-hospital 24-hour blood pressure monitoring in patients with decompensated atrial fibrillation (AF) revealed that patients with a dipper blood pressure pattern exhibited a favorable association with the change of NT-proBNP levels.3 Patients with coronary slow flow phenomenon with scar tissue on cardiac magnetic resonance imaging have higher NT-proBNP levels.4 In addition, several studies have noted that NT-proBNP contributes to the incidence of coronary artery disease by increasing the risk of heart failure disease and is an important assessment indicator of coronary artery disease.5-
The collective microbial community of all microorganisms in the human gut is termed “gut microbiota (GM).”6 The host provides the environment and nutrients necessary for GM survival, and GM actively participates in various physiological processes within the human body in turn. In recent years, studies have found a close relationship between GM and heart failure. Some researchers have detected identical bacterial DNA in arterial plaques and the intestinal flora of coronary heart disease (CHD) patients, suggesting that GM may be a potential source of bacteria in arterial plaques, and it may be involved in the development of CHD.7 Although heart failure contributes to cardiovascular diseases such as CHD8 and myocardial infarction,9 and GM is associated with heart failure-induced cardiovascular disease,10 the causality of GM and heart failure remains debated. Modulating or remodeling the GM promises to be a new clinical therapeutic idea for the effective and specific treatment of heart failure.
Mendelian randomization is a statistical method that emphasizes overcoming confounders and reverse causality bias. This approach is based on the fact that IVs are associated with the risk factor or biomarker being studied but not with other potential confounders. Thus, these instrumental variables (IVs) can be considered randomly assigned to individuals, similar to random assignment in an experiment. By adopting this approach, causality can be assessed more reliably without interference from other factors, increasing the credibility of study findings.11 Although randomized controlled trials (RCTs) are the gold standard for evaluating the safety and efficacy of an intervention, they are challenging to implement because of the complexity of their design, the high cost in terms of time, human and material resources, and ethical constraints. Conceptually, MR is similar to RCTs in that genetic variables are randomly assigned at birth to “case” or “control” groups as IVs and remain constant throughout the life cycle, according to Mendel’s second law. Genome-wide association study (GWAS) has played an essential role in complex disease genetics by searching for genotype-phenotype relationships through millions of genetic variations tested on the genomes of many individuals.12 Single nucleotide polymorphism (SNP) exposure and SNP-outcome associations can be extracted separately from independent GWAS and used in MR analyses of 2 independent samples to generate a single causal estimate. Thus, these enabled us to infer a causal relationship between GM and NT-proBNP using a Mendelian randomization (MR) approach. It is independent of confounders between genetic variation and outcome, and serves as a novel approach to studying causal relationships between GM and NT-proBNP.13-
Methods
Study Design
Intestinal flora and NT-proBNP were respectively used as the exposure factor and outcome variable, with significantly associated SNPs serving as IVs. Based on publicly available GWAS summary statistics, the causal relationship between 119 bacterial taxa and NT-proBNP was assessed using 2-sample bidirectional MR analysis. This study adhered to 3 crucial assumptions of the MR methodology: (1) there is a significant association between IVs and gut flora; (2) IVs are not associated with any of the gut flora-NT-proBNP confounders; and (3) IVs affect NT-proBNP only through the gut flora, instead of any other pathway.
Data Source
Genetic data on gut flora were obtained from the GWAS meta-analysis published by the largest MiBioGen consortium, which collected 16S rRNA gene sequencing profiles and genotyping data from 18 340 subjects (13 266 of European origin) from 11 countries, spanning Europe and the United States, and analyzed the gut microbiome signature loci.16 In this study, bacterial taxa were analyzed at the genus level, encompassing 119 bacterial taxa after excluding 15, and a total of 3301 NT-proBNP sources, all of European origin,17 were included. The NT-proBNP was from a population with a rigorously collected set of demographic data, anthropometric measurements, lifestyle factors, and dietary information. To ensure a clean baseline for outcome assessment, participants with a history of major diseases, recent illnesses, or infections were excluded from the study. The NT-proBNP levels were measured using SomaLogic assays, which employ SOMAmer probes to accurately identify and bind target proteins. This advanced method allows for the detection of proteins even at low concentrations, providing precise and reliable measurements essential for evaluating NT-proBNP levels. The NT-proBNP datasets were chosen primarily from European ancestry populations, consistent with the fact that the GM exposure data, which also predominantly comes from European ancestry. This was done to maintain consistency and reduce confounding variables related to genetic and environmental diversity.
Instrumental Variable
First, SNPs with significant association with GM (
Mendelian Randomization Analysis
In this study, 5 methods were used to verify the causal relationship between GM and NT-proBNP in 119 taxa: MR-random or fixed effects inverse variance weighted (IVW), weighted median estimation (WME), MR-Egger regression, simple mode (SM), and weighted mode (WM). For features containing only 1 IV, the Wald ratio test was used to estimate the association between the identified IV and NT-proBNP. Results with multiple IVs were mainly based on the IVW method, supplemented by the other 4 methods.
Sensitivity Analysis
To ensure the reliability of the causal effect assessment results, sensitivity analysis was conducted in this study. The leave-one-out tests were performed to examine the effect of individual IVs on the overall results. A test for heterogeneity was performed using Cochran’s Q statistics to assess whether there were differences between IVs in the 2-sample MR analysis. Q statistics significant at a
Statistical Analysis
Data were organized and analyzed using R software (version 4.2.0). The “
Results
Instrumental Variables Selection
Based on the IVW method, a total of 9 out of 104 GM classes were found to be causally associated with NT-proBNP (
Mendelian Randomization Analysis of Gut Microbiota and N-Terminal Pro-B-Type Natriuretic Peptide
As shown in
Consistent with the above results, the overall direction of causality between GM and NT-proBNP calculated by the 5 methods is shown in
Sensitivity Analysis Results
The test for heterogeneity of the gut microorganisms of the 9 taxa showed
Discussion
Based on MR analysis, a causal association was found between 9 intestinal bacterial species and NT-proBNP. Among them, genera
It is known that BNP is expressed by the myocardium in response to elevated atrial wall pressure, both BNP as well as NT-proBNP have become important biomarkers of cardiovascular disease.20 Quantification of NT-proBNP has been shown to predict the progression of coronary artery disease. One previous study had found different doses of azithromycin affected the abundance and grouping of GM in rat models of heart failure, and the degree of myocardial damage, GM variations, and clinical manifestations of heart failure were found to be consistent with each other.21 Another study noted that hypertension could result in the imbalance of GM, while chronic hypertension could lead to dysregulation of fecal microbial ecology and elevated levels of the biomarker NT-proBNP.22 Although the above studies indicate that gut flora may be associated with NT-proBNP-induced heart failure disease, no studies have directly demonstrated a causal relationship between GM and NT-proBNP.
In the present study, it was found that the genus
It was found that the genera, UCG 008, group, group, and group were increased factors for NT-proBNP. The genera
The NT-proBNP is an essential biomarker for diagnosing heart failure in clinical practice. The level of NT-proBNP increases with the severity of heart failure, and chronic heart failure patients often have intestinal dysfunction.32 The GM can regulate and maintain intestinal and overall host health by releasing secretions. However, there is currently no direct evidence to suggest the mechanism by which GM regulates NT pro-BNP in the research findings. Studies have shown that GM products short-chain fatty acid (SCFA) can promote intestinal fermentation processes, which is beneficial for the prognosis of heart failure patients.33 Whereas in the results, genus
Although it has been suggested that the GM would be a potential therapeutic target for heart failure as it plays a key role in regulating host physiology and metabolism and in the development of heart failure. However, the discovery of 9 GMs with a causal relationship with heart failure has not been applied in the clinical setting. Only studies have used genera
This study used large-scale GWAS data and MR methods to analyze the causal relationship between 104 GM classes and NT-proBNP. The present study, due to its large sample size, helped to reduce sampling error and was able to help exclude the effect of confounding factors, resulting in a more accurate assessment of the causal relationship between GM and NT-proBNP. In addition, the present study conducted several sensitivity analyses to provide comprehensive quality control on aspects such as multiplicity and heterogeneity of MR analyses, and thus, the conclusions obtained are robust.
The present study provides the most comprehensive assessment of the causal relationship between GM and NT-proBNP based on MR methods, but this study has some limitations. First, due to the limited current understanding of the etiology and mechanisms of abnormal NT-proBNP levels in humans, if genetic tools influence NT-proBNP through confounding variables other than exposure variables, it may introduce observational bias, leading to inaccurate causal estimates. Based on these potential confounding variables, an inherent limitation of the MR method, the results will also be validated using RCTs in the future. Second, there may be gene-environment interactions in the effect of SNPs on exposure factors, implying that SNPs may have a nonlinear impact on the risk of NT-proBNP abnormalities; however, MR analyses can only assess linear associations and cannot evaluate the effect of extremes of exposure factors on disease. In addition, the results of the MR analyses in this study may not be generalizable to populations outside of European ancestry or populations residing in different geographic regions, as genetic heterogeneity and environmental factors vary between areas. This data was not analyzed for specific stratification by gender, age, dietary habits, etc. Finally, complex physiological mechanisms are involved in causing NT-proBNP abnormalities, and the MR analyses performed in this study may not fully capture this causal relationship. Therefore, if GM is to be regulated as a clinical measure to combat diseases such as heart failure caused by NT-proBNP abnormalities, it must be refined by completing higher-quality MR analyses or RCTs. In addition, this study could not point out the specific mechanisms by which GM exerts its effects on NT-proBNP, and further studies are needed to reveal the pathophysiological mechanisms underlying these causal relationships.
Conclusion
In summary, this study explored the causal relationship between GM and NT-proBNP applying MR analysis. Of note, this study indicates that regulating gut flora may potentially prevent and control NT-proBNP abnormalities, thereby reducing the risk of NT-proBNP-related heart failure and other diseases, which lays the foundation for further exploring the mechanism linking GM with heart failure and other diseases. It is also acknowledged that the study’s focus on populations of European ancestry might limit the generalizability of the findings to other populations.
Footnotes
References
- Averina M, Stylidis M, Brox J, Schirmer H. NT-ProBNP and high-sensitivity troponin T as screening tests for subclinical chronic heart failure in a general population. ESC Heart Fail. 2022;9(3):1954-1962.
- Felker GM, Anstrom KJ, Adams KF. Effect of natriuretic peptide-guided therapy on hospitalization or cardiovascular mortality in high-risk patients with heart failure and reduced ejection fraction: a randomized clinical trial. JAMA. 2017;318(8):713-720.
- Candemir M, Kızıltunç E, Nurkoç SG, Cihan B, Şahinarslan A. Predictors of length of hospital stay and in-hospital adverse events in patients with acute decompensated heart failure: in-hospital 24-hour blood pressure monitoring data. Hellenic J Cardiol. ;():-.
- Candemir M, Şahinarslan A, Yazol M, Öner YA, Boyacı B. Determination of myocardial scar tissue in coronary slow flow phenomenon and the relationship between amount of scar tissue and Nt-ProBNP. Determinação do Tecido Cicatricial do Miocárdio no Fenômeno de Fluxo Coronário Lento e a Relação entre a Quantidade de Tecido Cicatricial e o Nt-ProBNP. Arq Bras Cardiol. 2020;114(3):540-551.
- Sterling MR, Durant RW, Bryan J. N-terminal pro-B-type natriuretic peptide and microsize myocardial infarction risk in the reasons for geographic and racial differences in stroke study. BMC Cardiovasc Disord. 2018;18(1):66-.
- Vemuri R, Gundamaraju R, Shastri MD. Gut microbial changes, interactions, and their implications on human lifecycle: an ageing perspective. BioMed Res Int. 2018;2018():4178607-.
- Guo Y, Li H, Liu Z. Impaired intestinal barrier function in a mouse model of hyperuricemia. Mol Med Rep. 2019;20(4):3292-3300.
- Yang Z, Ma H, Yin D, Sun C. Impact of Sacubitril/Valsartan on cardiac structure and blood levels of miRNA-328 and NT-proBNP in patients with CHD and chronic heart failure. Altern Ther Health Med. ;():-.
- Jering KS, Claggett BL, Pfeffer MA. Prognostic importance of NT-proBNP (N-terminal pro-B-type natriuretic peptide) following high-risk myocardial infarction in the PARADISE-MI trial. Circ Heart Fail. 2023;16(5):e010259-.
- Fan Y, Liang L, Tang X. Changes in the gut microbiota structure and function in rats with doxorubicin-induced heart failure. Front Cell Infect Microbiol. 2023;13():1135428-.
- Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318(19):1925-1926.
- Visscher PM, Wray NR, Zhang Q. 10 years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5-22.
- Dan YL, Wang P, Cheng Z. Circulating adiponectin levels and systemic lupus erythematosus: a two-sample Mendelian randomization study. Rheumatol (Oxf Engl). 2021;60(2):940-946.
- Long Y, Tang L, Zhou Y, Zhao S, Zhu H. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. 2023;21(1):66-.
- Sanna S, van Zuydam NR, Mahajan A. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51(4):600-605.
- Kurilshikov A, Medina-Gomez C, Bacigalupe R. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53(2):156-165.
- Sun BB, Maranville JC, Peters JE. Genomic atlas of the human plasma proteome. Nature. 2018;558(7708):73-79.
- Shen H, He Q, Shao X. Predictive value of NT-proBNP and hs-TnT for outcomes after pediatric congenital cardiac surgery. Int J Surg. 2024;110(6):3365-3372.
- Zeng Q, Li D, He Y. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci Rep. 2019;9(1):13424-.
- de Lemos JA, McGuire DK, Drazner MH. B-type natriuretic peptide in cardiovascular disease. Lancet (London, England). 2003;362(9380):316-322.
- Vemuri R, Ruggiero A, Whitfield JM. Hypertension promotes microbial translocation and dysbiotic shifts in the fecal microbiome of nonhuman primates. Am J Physiol Heart Circ Physiol. 2022;322(3):H474-H485.
- Wang L, Zhou W, Guo M. The gut microbiota is associated with clinical response to statin treatment in patients with coronary artery disease. Atherosclerosis. 2021;325():16-23.
- Yang T, Qu H, Song X. Luhong granules prevent ventricular remodelling after myocardial infarction by reducing the metabolites TMAO and LPS of the intestinal flora. Evid Based Complement Alternat Med. 2019;2019():8937427-.
- Zuo K, Li J, Li K. Disordered gut microbiota and alterations in metabolic patterns are associated with atrial fibrillation. GigaScience. 2019;8(6):giz058-.
- Tian Y, Yao G, Skudder-Hill L. Gut microbiota’s causative relationship with peripheral artery disease: a Mendelian randomization study. Front Microbiol. 2024;15():1340262-.
- Mao M, Zhai C, Qian G. Gut microbiome relationship with arrhythmias and conduction blocks: a two-sample Mendelian randomization study. J Electrocardiol. 2023;80():155-161.
- Jiao J, Zhang Y, Han P, Zhai S. A preliminary study on the value of intestinal flora in predicting major adverse cardiovascular and cerebrovascular events in patients with refractory hypertension. Comp Math Methods Med. 2022;2022():7723105-.
- Zhu Q, Gao R, Zhang Y. Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol Genomics. 2018;50(10):893-903.
- Zhu S, Gao B, Umetsu Y. Adaptively evolved human oral actinomyces-sourced defensins show therapeutic potential. EMBO Mol Med. 2022;14(2):e14499-.
- Liu J, An N, Ma C. Correlation analysis of intestinal flora with hypertension. Exp Ther Med. 2018;16(3):2325-2330.
- Han Y, Gong Z, Sun G. Dysbiosis of gut microbiota in patients with acute myocardial infarction. Front Microbiol. 2021;12():680101-.
- Otto CM, Rahimi K. Heartbeat: the gut microbiota and heart failure. Heart (Br Card Soc). 2016;102(11):811-812.
- Modrego J, Ortega-Hernández A, Goirigolzarri J. Gut microbiota and derived short-chain fatty acids are linked to evolution of heart failure patients. Int J Mol Sci. 2023;24(18):13892-.
- van der Wal HH, Comin-Colet J, Klip IT. Vitamin B12 and folate deficiency in chronic heart failure. Heart (Br Card Soc). 2015;101(4):302-310.
- Hu W, Yuan L, Wang X. Predictive value of arterial blood lactic acid concentration on the risk of in-hospital all-cause death in patients with acute heart failure. Int J Clin Pract. 2022;2022():7644535-.
- Mayerhofer CCK, Ueland T, Broch K. Increased secondary/primary bile acid ratio in chronic heart failure. J Card Fail. 2017;23(9):666-671.
- Jie Z, Xia H, Zhong SL. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun. 2017;8(1):845-.
- Bartolomaeus H, Balogh A, Yakoub M. Short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation. 2019;139(11):1407-1421.
- Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12α-hydroxylated bile acids. Diabetes. 2013;62(12):4184-4191.
- Rahman MM, Islam F, -Or-Rashid MH. The gut microbiota (microbiome) in cardiovascular disease and its therapeutic regulation. Front Cell Infect Microbiol. 2022;12():903570-.


